All living cells contain proteins with different functions, depending on the type of cell.
Researchers at the University of Gothenburg in Sweden have discovered a way to identify proteins even without looking at their structure. They say their method is faster, easier and more reliable than previous methods.
Currently, the general view is that each protein’s structure is what controls its function in cells. The atomic sequences, meaning how the atoms are arranged in the proteins, create the protein’s structure and shape. But there are many proteins that lack a well-defined structure.
Researcher Gergely Katona has developed a new method where proteins are scanned based on the number of amino acids (or the number of different atoms) they contain in order to identify them and their function instead of identifying them based on their structure. With this scanning method, the researchers were able to predict relatively reliably, which combination of amino acids is needed to bind to the protein survivin. The outcome was a reliability of about 80%, which is better than when the protein’s primary structures are used for identification. The results were published in the scientific journal iScience.
The structure of less importance
Several thousand peptides containing 15 amino acids were tested and the researchers were able to conclude that it was the amino acid content that affected their binding to survivin, while the structure of the peptides had almost no significance.
“Simply counting things has often been a successful method in science. Here, we counted the number of amino acids and were able to predict the function of the protein surprisingly well,” Katona said.
The researchers said they see advantages with this method of scanning proteins. Machine learning/artificial intelligence also speeds up the process of linking the number and type of amino acids to a certain function. This in turn, means that the development of new biological drugs can be accelerated.
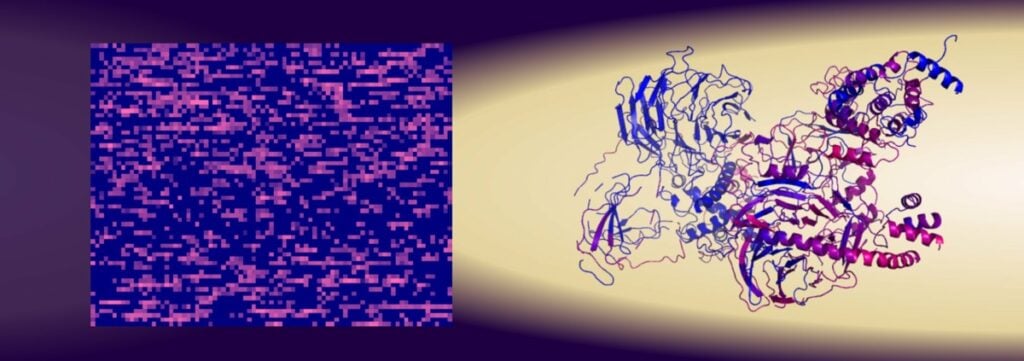
In the researchers’ experiments with this new scanning method, a completely new function of the protein survivin was also discovered. This protein is mainly prominent in embryo cells and prevents programmed cell death. But in cancerous tumors, survivin becomes unregulated and thus facilitates the development of the cancer.
Useful in cancer research
The researchers have now seen that survivin directly influences another protein, PRC2, which switches off and on various functions in the DNA in the cell, like a kind of programming. Dysfunctional PRC2 can also be linked to various forms of cancer. Today’s cancer drugs target both survivin and PRC2, but with the newly discovered link between survivin and PRC2, the drugs may need to be designed differently to avoid serious side effects.
“We saw that if we suppressed the level of survivin, activity in PRC2 increased. The dream for pharmaceutical companies is to find the right targets in the atomic sequences to be able to balance the two proteins,” Katona concluded.

